Mission of the Core
The mission of the Biospecimens Core is to obtain high-quality, carefully annotated biospecimens in sufficient quantity to enable investigators to tackle highly significant scientific questions. To accomplish this goal, the Core provides the following services:
- Collect, process, bank and distribute specimens from patients and family members who have consented to participate in the Iowa NET Registry. These specimens include DNA from patients with NETs and family members, excess fresh tumor and adjacent normal tissue obtained at surgery, and associated paraffin blocks
- Acquire, process, and distribute de-identified NET specimens obtained through the Iowa Residual Tumor and Virtual Tissue Repositories. Tumor/normal tissue blocks, paired with Cancer Registry data are available.
- Provide accurate tumor classification according to the 2017 WHO Classification of Tumours of Endocrine Organs for tumor tissue collected from Registry patients or obtained from repositories. Tumor specimens lacking pathology report or other information vital to complete classification are analyzed for verification of diagnosis and grade. This is particularly important for specimens in which the diagnosis was made prior to publication of the 2010 WHO Classification.
- Maintain data obtained from Iowa NET Registry biospecimens and SEER Residual Tissue Repository biospecimens in secure databases.
Core and Projects Collaborations
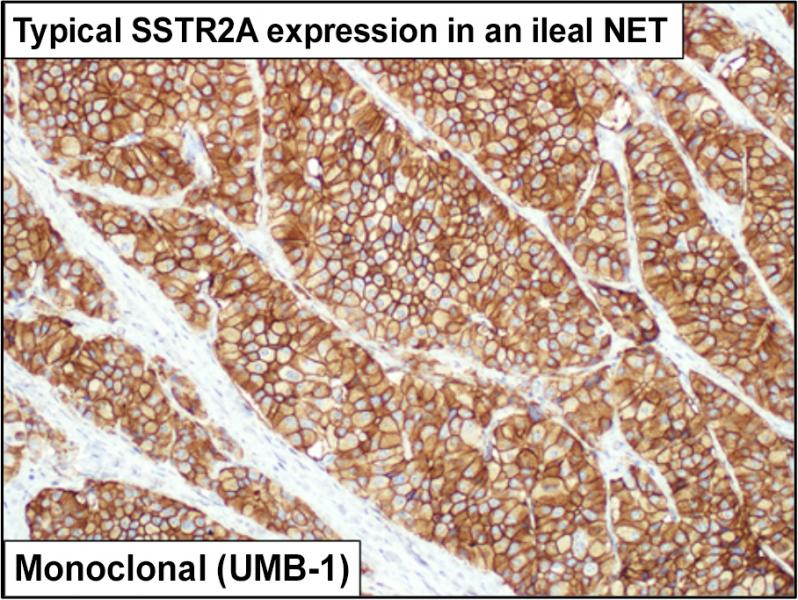
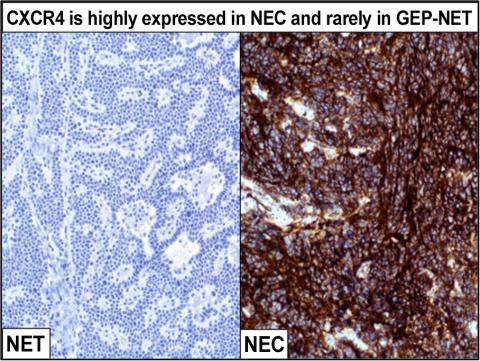
The core is highly interested in novel nuclear medicine imaging and peptide receptor radionuclide therapy (PRRT) targets. Dr. Bellizzi, an expert in diagnostic immunohistochemistry (IHC), has developed IHC tests for SSTR2A and CXCR4, two key PRRT targets. The Core has stained and evaluated archival slides from hundreds of Iowa Neuroendocrine Tumor Registry patients for SSTR2A and has maintained a prospective database of CXCR4-expressing neuroendocrine carcinomas to identify patients for [68Ga]DOTATOC- and [68Ga]Pentixafor-PET/CT.
The core has collaborated extensively with Dr. Quelle (Project 2) in developing immunohistochemical markers of signaling pathways regulating cell cycle progression and applying these to cell lines subjected to various experimental conditions, as well as tissue microarrays (TMAs) of human tumors. The core has also collaborated with Dr. Darbro (Project 2) in the validation of novel diagnostic fluorescence in situ hybridization (FISH) targets.
The Core has worked with Dr. Howe (Project 3) to develop a large set of TMAs representing surgically resected tumors. This includes over 600 unique tumors, including primary, regional, and distant disease. The core is well-positioned to use immunohistochemistry to validate markers, either diagnostic or prognostic, discovered through gene expression profiling.

Clinical Research on Neuroendocrine Tumors
Neuroendocrine epithelial neoplasms include well-differentiated neuroendocrine tumors and poorly differentiated neuroendocrine carcinomas (i.e., small cell and large cell neuroendocrine carcinoma). Dr. Bellizzi has a long-standing interest and expertise in the diagnosis and classification of neuroendocrine epithelial neoplasms.
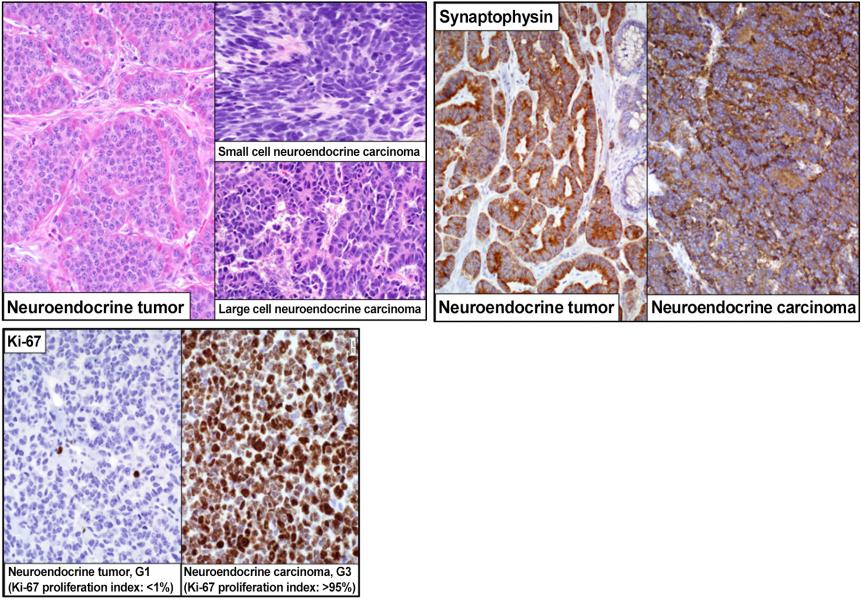
The well-differentiated neuroendocrine tumors are graded (G1, G2, G3) based on mitotic count and Ki-67 immunohistochemistry. Poorly differentiated neuroendocrine carcinomas are G3, by definition. The grade has prognostic and therapeutic implications. Dr. Bellizzi has validated numerous immunohistochemical stains that serve various diagnostic and predictive roles in neuroendocrine tumors.
| Marker | Clinical Application |
|---|---|
| SSTR2A | Highly expressed by NETs and some NECs; correlates with results of SRI; predicts response to PRRT; prognostically favorable |
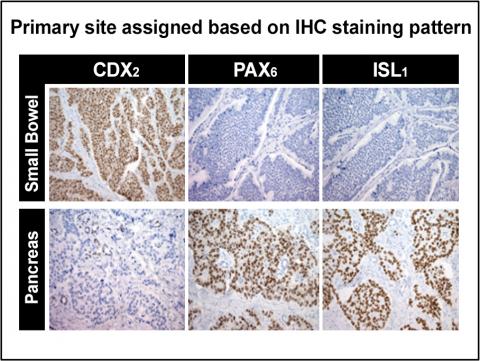
| PAX6 | Marker of pancreatic origin in NET |
| Islet 1 | Marker of pancreatic origin in NET |
| SDHB | Absence supports a diagnosis of paraganglioma/pheochromocytoma over NET; absence typical of patients with germline SDH subunit mutations |
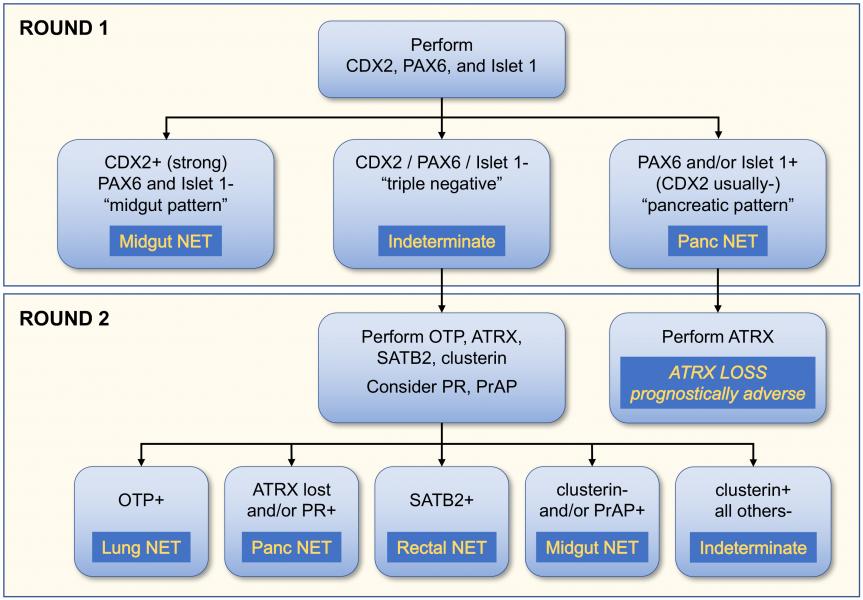
| ATRX | Absence supports a diagnosis of pancreatic NET; absence is prognostically adverse |
| SATB2 | Marker of rectal NET |
| OTP | Marker of bronchopulmonary NET; absence in a bronchopulmonary NET is prognostically adverse |
| Clusterin | Absence in a NET supports an ileal origin; absence in a G3 tumor supports a diagnosis of NEC |
| CXCR4 | Highly expressed by NECs and a limited subset of NETs; correlates with results of Pentixafor scan; predicts response to PRRT; prognostically adverse |
| Rb | Absence is a genetic hallmark of NEC |
| Serotonin | Marker of midgut origin in NET |
Dr. Bellizzi is also currently optimizing a classifier to assign the site of origin of a metastatic well-differentiated neuroendocrine tumor of occult origin. 15-20% of NETs present in this fashion and determining the site of origin is prognostically and therapeutically significant.
The primary classifier includes the midgut marker CDX2 and the pancreatic markers PAX6 and Islet 1. When this initial panel is negative, additional immunostains are employed.
Another important aspect of pathological diagnosis is the distinction of well-differentiated NET, G3 from large cell neuroendocrine carcinoma (LCNEC). ATRX, clusterin, and Rb have recently been validated to serve this goal:
Supportive of a Diagnosis of NET, G3
- P53 wild-type pattern
- Rb intact
- Clusterin expressed
- CDX2 expressed (midgut origin)
- PAX6 and/or ISL1 expressed (pancreatic origin)
- ATRX absent (pancreatic origin)
Supportive of a Diagnosis of LCNEC
- p53 missense or null pattern
- Rb absent
- Clusterin not expressed
The final diagnosis, including tissue of origin, tumor Grade 1, 2, or 3, and NET vs NEC is extremely important for determination of optimal treatment of the patient and is also a key to accurate and reproducible scientific research.
Core Leaders

Andrew Bellizzi, MD
Clinical Professor of Pathology
Email: andrew-bellizzi@uiowa.edu
