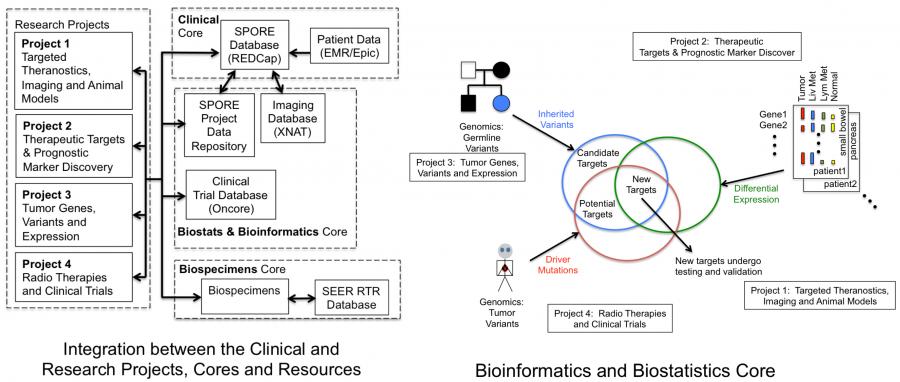
Robust Biostatistics and Bioinformatics integration is critical for Iowa Neuroendocrine Tumor SPORE investigators to conduct effective translational research in neuroendocrine tumors. The Biostatistics and Bioinformatics Core provides this expertise through full integration of Biostatistics and Bioinformatics into project design, implementation, execution, analysis and publication; through synergistic facilitation of shared research outcomes among the various project groups of the SPORE; and through career development and clinical investigator training.
The Biostatistics and Bioinformatics Core provides statistical design, collaborative analysis, and data management support for each of the Iowa NET SPORE projects, as well as for developmental and career enhancement projects. Biostatistics and Bioinformatics Core seeks to build a synergistic interaction with investigators through data gatekeeping and with all project groups through data analyses, information sharing, data form development and processing, data collection and entry, data archiving, quality control, and clinical trial design and data analysis.
Specific Aims
Aim 1
Provide integrative statistical and bioinformatics support across different fields, including basic science, translational and clinical trials. Core directors work closely with investigators to develop statistical design and analysis plans for both clinical and laboratory projects. Data management for clinical trials through the Holden Comprehensive Cancer Center (HCCC) OnCore data management system includes adverse event monitoring and oversight of the IRB-approved Neuroendocrine Registry in REDCap. In collaboration with the Biospecimens Core, we will track acquisition and use of all identified tissues, including exome sequencing of tumor and normal tissues, as well as G protein coupled receptor, oncogene, and tumor suppressor gene expression in tumors. Similarly, collaboration with the Clinical Research Core will enable the Biostatistics and Bioinformatics Core to integrate individual subject participation in imaging and therapeutic trials, their response to therapies and resultant quality of life information as well as long-term follow-up into the Iowa Neuroendocrine Tumor Database.
Aim 2
Provide Bioinformatics support and analysis critical for translational projects aimed at identification of gene mutations, deletions, and tumor initiating events in neuroendocrine tumors. Areas of support will include analysis of next-generation sequencing results, chromosomal integrity, and gene expression arrays. The Bioinformatics Core has established a cancer genome variant calling analysis pipeline. Briefly, whole-exome reads are mapped. Both germline and somatic variants (mutations) are identified using peer-reviewed best-practices and workflows to ensure robust and unbiased results. Additional parameters include the segregation pattern of disease in families and variant pathogenicity prediction. Scientific rigor is emphasized by the use of the American College of Medical Genetics and Genomics (ACMG) guidelines within a structured framework of evidence-based codes by which to score a variant.
RNA-sequencing analysis enables investigation of gene expression differences between primary tumor, liver metastases, lymph metastases and normal tissue for small bowel and pancreas NETs. Rigor is obtained through the use of appropriate statistical methods implemented by community-accepted tools for quantitation and mapping. The study design includes rigor through the use of replicates, and significantly differentially expressed genes are determined in an unbiased way with respect to sample type (cancer vs normal).
Aim 3
Provide a mechanism for the management and integration of existing and newly collected data in basic and translational projects. This aim will be accomplished through consistent and compatible data handling, database development, data form development and processing, data collection and entry, data archiving, quality control, and management of information relating to gene mutation identification and genotyping as well as gene expression data for integration into translational, population, and clinical correlative studies.
Innovative Clinical trial Designs for Dual-Target, Dual-Radionuclide Therapy.
Each individual subject receives dosimetry-guided, optimal dosing of 90Y-DOTATOC & 131I-MIBG or 177Lu-DOTATATE & 131I-MIBG. Dosimetry guides the amount of each radiopharmaceutical in these unique combinations that will increase radiation dose to tumor while maintaining a safe dose to bone marrow and kidneys.
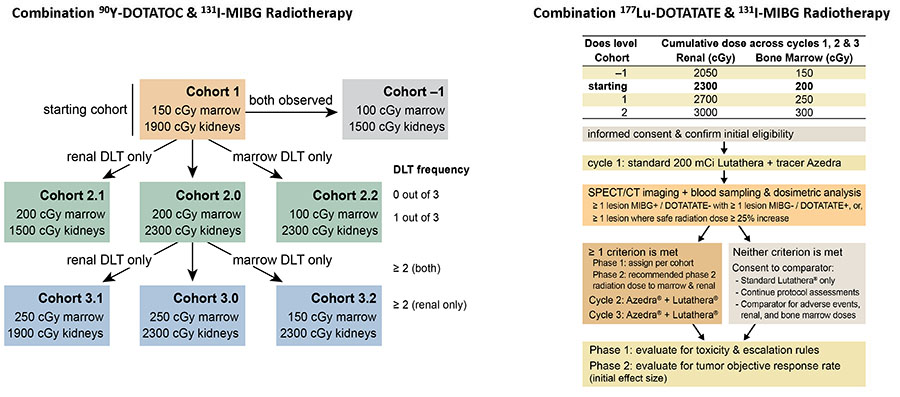
DLT: dose limiting toxicity; Lutathera: 177Lu-DOTATATE; Azedra: 131I-MIBG